Tutorial
Basic example:
Modeling lactate dehydrogenase from Trichomonas vaginalis based on a
single template
All input and output files for this example are available to download,
in either zip format (for Windows) or
.tar.gz format (for Unix/Linux).
A novel gene for lactate dehydrogenase was identified from the genomic
sequence of Trichomonas vaginalis (TvLDH). The corresponding protein
had a higher similarity to the malate dehydrogenase of the same species (TvMDH)
than to any other LDH. We hypothesized that TvLDH arose from TvMDH by
convergent evolution relatively recently. Comparative models were constructed
for TvLDH and TvMDH to study the sequences in the structural context and to
suggest site-directed mutagenesis experiments for elucidating specificity
changes in this apparent case of convergent evolution of enzymatic specificity.
The native and mutated enzymes were expressed and their activities were
compared.
The individual modeling steps of this example are explained below. Note
that we go through every step in this tutorial to build a model knowing only
the amino acid sequence. In practice you may already know the related
structures, and may even have an alignment from another program, so you can
skip one or more steps. Alternatively, for very simple applications you
may be able to use the ModWeb web server rather than
Modeller itself.
1. Searching for structures related to TvLDH
First, it is necessary to put the target TvLDH sequence into the PIR format
readable by MODELLER (file
`TvLDH.ali').
>P1;TvLDH
sequence:TvLDH:::::::0.00: 0.00
MSEAAHVLITGAAGQIGYILSHWIASGELYGDRQVYLHLLDIPPAMNRLTALTMELEDCAFPHLAGFVATTDPKA
AFKDIDCAFLVASMPLKPGQVRADLISSNSVIFKNTGEYLSKWAKPSVKVLVIGNPDNTNCEIAMLHAKNLKPEN
FSSLSMLDQNRAYYEVASKLGVDVKDVHDIIVWGNHGESMVADLTQATFTKEGKTQKVVDVLDHDYVFDTFFKKI
GHRAWDILEHRGFTSAASPTKAAIQHMKAWLFGTAPGEVLSMGIPVPEGNPYGIKPGVVFSFPCNVDKEGKIHVV
EGFKVNDWLREKLDFTEKDLFHEKEIALNHLAQGG*
File: TvLDH.ali
The first line contains the sequence code, in the format
`>P1;code'. The second line with ten fields separated by colons
generally contains information about the structure file, if applicable. Only
two of these fields are used for sequences, `sequence' (indicating
that the file contains a sequence without known structure) and `TvLDH'
(the model file name). The rest of the file contains the sequence of TvLDH,
with `*' marking its end. The standard one-letter amino acid codes are used.
(Note that they must be upper case; some lower case letters are used for
non-standard residues. See the file modlib/restyp.lib in the
Modeller distribution for more information.)
A search for potentially related sequences of known
structure can be performed by the SEQUENCE_SEARCH command of
MODELLER. The following script uses the query
sequence `TvLDH' assigned to the variable ALIGN_CODES
from the file `TvLDH.ali', which in turn is
assigned to the variable FILE (see file
`seq_search.top').
SET SEARCH_RANDOMIZATIONS = 100
SET FILE = 'TvLDH.ali'
READ_SEQUENCE_DB
SEQUENCE_SEARCH ALIGN_CODES = 'TvLDH', DATA_FILE = ON
File: seq_search.top
Modeller should be directed to run this script by using the following
command at your command line (be aware that it will take some time to run -
perhaps an hour or so - so be patient!):
mod7v7 seq_search.top
(You can get a command line using xterm or GNOME Terminal in Linux,
Terminal in Mac OS X, or Command Prompt in Windows. For more information on
running Modeller, see the manual.)
The SEQUENCE_SEARCH command has many options, but in this
example only SEARCH_RANDOMIZATIONS and
DATA_FILE are set to non-default values.
SEARCH_RANDOMIZATIONS specifies the number of times the query
sequence is randomized during the calculation of the significance score for
each sequence-sequence comparison. The higher the number of randomizations,
the more accurate the significance score. DATA_FILE = ON
triggers creation of an additional summary output file
(see `seqsearch.dat').
2. Selecting a template
The output of the `seq_search.top' script is
written to the `seq_search.log' file.
MODELLER always produces a log file. Errors and
warnings in log files can be found by searching for the `_E>' and
`_W>' strings, respectively. At the end of the log file,
MODELLER lists the hits sorted by alignment
significance. Because the log file is sometimes very long, a separate data file
is created that contains the summary of the search. The example shows only the
top 10 hits (file `seqsearch.dat').
(Note that your results may differ slightly from these, due to differences
in the search database and different floating-point precision on different
computers.)
1 TvLDH 1bdmA 335 318 139 41.5 43.7 202461. 36.1 -999.0 -999.0
2 TvLDH 1civA 335 374 115 30.7 34.3 195914. 24.7 -999.0 -999.0
3 TvLDH 1guzA 335 305 71 21.2 23.3 161076. 10.8 -999.0 -999.0
4 TvLDH 2cmd 335 312 78 23.3 25.0 161524. 8.9 -999.0 -999.0
5 TvLDH 1lldA 335 313 70 20.9 22.4 161130. 8.9 -999.0 -999.0
6 TvLDH 2hlpA 335 303 62 18.5 20.5 154717. 8.2 -999.0 -999.0
7 TvLDH 1ceqA 335 304 68 20.3 22.4 156953. 8.0 -999.0 -999.0
8 TvLDH 1hyeA 335 307 73 21.8 23.8 157429. 7.4 -999.0 -999.0
9 TvLDH 6ldh 335 329 60 17.9 18.2 161816. 6.0 -999.0 -999.0
10 TvLDH 1ez4A 335 307 61 18.2 19.9 155527. 5.9 -999.0 -999.0
File: seqsearch.dat
The most important columns in the SEQUENCE_SEARCH output
are the `CODE_2', `%ID' and `SIGNI' columns. The
`CODE_2' column reports the code of the PDB sequence that was compared
with the target sequence. The PDB code in each line is the representative of a
group of PDB sequences that share 40% or more sequence identity to each other
and have less than 30 residues or 30% sequence length difference. All the
members of the group can be found in the MODELLER
`CHAINS_3.0_40_XN.grp' file. The `%ID1'
and `%ID2' columns report the percentage sequence identities between
TvLDH and a PDB sequence normalized by their lengths, respectively. In
general, a `%ID' value above approximately 25% indicates a potential
template unless the alignment is short (i.e., less than 100 residues). A
better measure of the significance of the alignment is given by the
`SIGNI' column. A value above 6.0 is generally significant
irrespective of the sequence identity and length. In this example, one
protein family represented by 1bdm:A shows significant
similarity with the target sequence, at more than 40% sequence identity.
While some other hits are also significant, the differences between
1bdm:A and other top scoring hits are so pronounced
that we use only the first hit as the template. As expected,
1bdm:A is a malate dehydrogenase (from a thermophilic
bacterium). Other structures closely related to 1bdm:A
(and thus not scanned against by SEQUENCE_SEARCH) can be
extracted from the `CHAINS_3.0_40_XN.grp' file:
1b8v:A, 1bmd:A, 1b8u:A, 1b8p:A, 1bdm:A, 1bdm:B, 4mdh:A,
5mdh:A, 7mdh:A, 7mdh:B, and 7mdh:C. All these
proteins are malate dehydrogenases. During the project, all of them and other
malate and lactate dehydrogenase structures were compared and considered as
templates (there were 19 structures in total).However, for the sake of
illustration, we will investigate only four of the proteins that are
sequentially most similar to the target, 1bmd:A, 4mdh:A,
5mdh:A, and 7mdh:A. The following script
performs all pairwise comparisons among the selected proteins (file
`compare.top').
READ_ALIGNMENT FILE = '$(LIB)/CHAINS_all.seq',;
ALIGN_CODES = '1bmdA' '4mdhA' '5mdhA' '7mdhA'
MALIGN
MALIGN3D
COMPARE
ID_TABLE
DENDROGRAM
File: compare.top
The READ_ALIGNMENT command reads the protein sequences and
information about their PDB files. MALIGN calculates their
multiple sequence alignment, used as the starting point for the multiple
structure alignment. The MALIGN3D command performs an
iterative least-squares superposition of the four 3D structures.
COMPARE command compares the structures according to the
alignment constructed by MALIGN3D. It does not make an
alignment, but it calculates the RMS and DRMS deviations between atomic
positions and distances, differences between the mainchain and sidechain
dihedral angles, percentage sequence identities, and several other measures.
Finally, the ID_TABLE command writes a file with pairwise
sequence distances that can be used directly as the input to the
DENDROGRAM command (or the clustering programs in the
PHYLIP package). DENDROGRAM
calculates a clustering tree from the input matrix of pairwise distances,
which helps visualizing differences among the template candidates. Excerpts
from the log file are shown below (file
`compare.log').
Sequence identity comparison (ID_TABLE):
Diagonal ... number of residues;
Upper triangle ... number of identical residues;
Lower triangle ... % sequence identity, id/min(length).
1bmdA @14mdhA @25mdhA @27mdhA @2
1bmdA @1 327 168 168 159
4mdhA @2 51 333 328 137
5mdhA @2 51 98 333 138
7mdhA @2 49 41 41 351
.---------------------------------------------------- 1bmdA @1.9 49.0000
|
| .--- 4mdhA @2.5 2.0000
| |
.---------------------------------------------------------- 5mdhA @2.4 55.5000
|
.------------------------------------------------------------ 7mdhA @2.4
+----+----+----+----+----+----+----+----+----+----+----+----+
Excerpts of the file compare.log
The comparison above shows that 5mdh:A and
4mdh:A are almost identical, both sequentially and
structurally. They were solved at similar resolutions, 2.4 and 2.5Å,
respectively. However, 4mdh:A has a better
crystallographic R-factor (16.7 versus 20%), eliminating
5mdh:A. Inspection of the PDB file for
7mdh:A reveals that its crystallographic refinement
was based on 1bmd:A. In addition,
7mdh:A was refined at a lower resolution than
1bmdA (2.4 versus 1.9), eliminating 7mdh:A. These
observations leave only 1bmd:A and
4mdh:A as potential templates. Finally,
4mdh:A is selected because of the higher overall
sequence similarity to the target sequence.
3. Aligning TvLDF with the template
A good way of aligning the sequence of TvLDH with the structure of
4mdh:A is the ALIGN2D command in
MODELLER. Although ALIGN2D is
based on a dynamic programming algorithm, it is different from standard
sequence sequence alignment methods because it takes into account structural
information from the template when constructing an alignment. This task is
achieved through a variable gap penalty function that tends to place gaps in
solvent exposed and curved regions, outside secondary structure segments, and
between two positions that are close in space. As a result, the alignment
errors are reduced by approximately one third relative to those that occur
with standard sequence alignment techniques. This improvement becomes more
important as the similarity between the sequences decreases and the number
of gaps increases. In the current example, the template-target similarity
is so high that almost any alignment method with reasonable parameters will
result in the same alignment. The following MODELLER
script aligns the TvLDH sequence in file
`TvLDH.ali' with the
4mdh:A structure in the PDB file
`4mdh.pdb' (file
`align2d.top').
READ_MODEL FILE = '4mdh'
SEQUENCE_TO_ALI ALIGN_CODES = '4mdh'
READ_ALIGNMENT FILE = 'TvLDH.ali', ALIGN_CODES = ALIGN_CODES 'TvLDH', ADD_SEQUENCE = on
ALIGN2D
WRITE_ALIGNMENT FILE='TvLDH-4mdhA.ali', ALIGNMENT_FORMAT = 'PIR'
WRITE_ALIGNMENT FILE='TvLDH-4mdhA.pap', ALIGNMENT_FORMAT = 'PAP'
File: align2d.top
In the first line, MODELLER reads the
4mdh structure file. The
SEQUENCE_TO_ALI command transfers the sequence to the
alignment array and assigns it the name of `4mdh'
(ALIGN_CODES). The third line reads the TvLDH sequence from
file `TvLDH.seq', assigns it the name
`TvLDH' (ALIGN_CODES) and adds it to the alignment
array (`ADD_SEQUENCE = ON'). The fourth line executes the
ALIGN2D command to perform the alignment. Finally, the
alignment is written out in two formats, PIR
(`TvLDH-4mdhA.ali') and PAP
(`TvLDH-4mdhA.pap'). The PIR format is used by
MODELLER in the subsequent model building stage.
The PAP alignment format is easier to inspect visually. Due to the high
target-template similarity, there are only a few gaps in the alignment. In the
PAP format, all identical positions are marked with a `*' (file
`TvLDH-4mdhA.pap').
_aln.pos 10 20 30 40 50 60
4mdh -SEPIRVLVTGAAGQIAYSLLYSIGNGSVFGKDQPIILVLLDITPMMGVLDGVLMELQDCALPLLKDV
TvLDH MSEAAHVLITGAAGQIGYILSHWIASGELYG-DRQVYLHLLDIPPAMNRLTALTMELEDCAFPHLAGF
_consrvd ** ** ******* * * * * * * * **** * * * *** *** * *
_aln.p 70 80 90 100 110 120 130
4mdh IATDKEEIAFKDLDVAILVGSMPRRDGMERKDLLKANVKIFKCQGAALDKYAKKSVKVIVVGNPANTN
TvLDH VATTDPKAAFKDIDCAFLVASMPLKPGQVRADLISSNSVIFKNTGEYLSKWAKPSVKVLVIGNPDNTN
_consrvd ** **** * * ** *** * * ** * *** * * * ** **** * *** ***
_aln.pos 140 150 160 170 180 190 200
4mdh CLTASKSAPSIPKENFSCLTRLDHNRAKAQIALKLGVTSDDVKNVIIWGNHSSTQYPDVNHAKVKLQA
TvLDH CEIAMLHAKNLKPENFSSLSMLDQNRAYYEVASKLGVDVKDVHDIIVWGNHGESMVADLTQATFTKEG
_consrvd * * * **** * ** *** * **** ** * **** * *
_aln.pos 210 220 230 240 250 260 270
4mdh KEVGVYEAVKDDSWLKGEFITTVQQRGAAVIKARKLSSAMSAAKAICDHVRDIWFGTPEGEFVSMGII
TvLDH KTQKVVDVLDHD-YVFDTFFKKIGHRAWDILEHRGFTSAASPTKAAIQHMKAWLFGTAPGEVLSMGIP
_consrvd * * * * * * ** * ** * *** ** ****
_aln.pos 280 290 300 310 320 330
4mdh SD-GNSYGVPDDLLYSFPVTIK-DKTWKIVEGLPINDFSREKMDLTAKELAEEKETAFEFLSSA-
TvLDH VPEGNPYGIKPGVVFSFPCNVDKEGKIHVVEGFKVNDWLREKLDFTEKDLFHEKEIALNHLAQGG
_consrvd ** ** *** *** ** *** * * * * *** * *
File: TvLDH-4mdhA.pap
4. Model building
Once a target-template alignment is constructed,
MODELLER calculates a 3D
model of the target completely automaticaly. The following script will
generate five similar models of TvLDH based on the
4mdh template structure and the alignment in file
`TvLDH-4mdhA.ali' (file
`model-single.top').
INCLUDE
SET ALNFILE = 'TvLDH-4mdhA.ali'
SET KNOWNS = '4mdh'
SET SEQUENCE = 'TvLDH'
SET STARTING_MODEL = 1
SET ENDING_MODEL = 5
CALL ROUTINE = 'model'
File: model-single.top
The first line includes many standard variable and routine definitions. The
following five lines set parameter values for the `model' routine.
ALNFILE names the file that contains the target-template
alignment in the PIR format. KNOWNS defines the known template
structure(s) in ALNFILE
(`TvLDH-4mdh.ali'). SEQUENCE
defines the name of the target sequence in ALNFILE.
STARTING_MODEL and ENDING_MODEL define the
number of models that are calculated (their indices will run from 1 to 5).
The last line in the file calls the `model' routine that
actually calculates the models. The most important output files are
`model-single.log', which reports warnings,
errors and other useful information including the input restraints used for
modeling that remain violated in the final model, and
`TvLDH.B99990001.pdb', which contains the model
coordinates in the PDB format. The model can be viewed by any program that
reads the PDB format, such as Chimera.
5. Model evaluation
If several models are calculated for the same target, the ``best'' model
can be selected by picking the model with the lowest value of the
MODELLER objective function, which is reported in
the second line of the model PDB file. The value of the objective function in
MODELLER is not an absolute measure in the sense
that it can only be used to rank models calculated from the same alignment.
Once a final model is selected, there are many ways to assess it. In this
example, PROSAII is used to evaluate the model
fold and PROCHECK is used to
check the model's stereochemistry. Before any external evaluation of the
model, one should check the log file from the modeling run for runtime errors
(`model-single.log') and restraint violations
(see the MODELLER
manual for details).
Both PROSAII and
PROCHECK confirm that a reasonable model was
obtained, with a Z-score comparable to that of the template (-10.53 and
-12.69 for the model and the template, respectively). However, the
PROSAII energy profile indicates a possible error
in the long active site loop between residues 90 and 100.
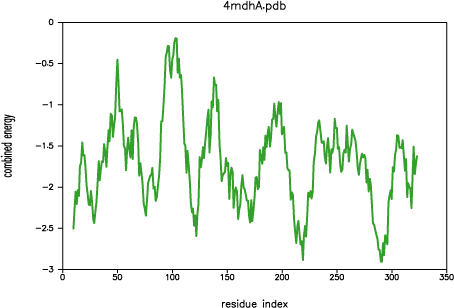
PROSAII profile for template 4mdh:A
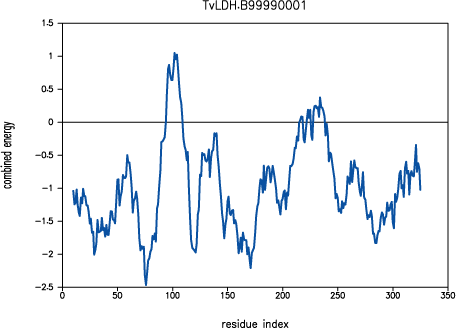
PROSAII profile for model TvLDH.B99990001
This loop interacts with region 220-250, that forms the other half of the
active site. This latter part is well resolved in the template and probably
correctly modeled in the target structure, but due to the unfavourable
non-bonded interactions with the 90-100 region, it is also reported to be
in error by PROSAII. In general, an error indicated
by PROSAII is not neccessarily an actual error,
especially if it highlights an active site or a protein-protein interface.
However, in this case, the same active site loops have a better profile
in the template structure, which strengthens the assessment that the model
is probably incorrect in the active site region. This problem is addressed
in the advanced modeling tutorial.
|