Modeling with cryo-EM
Step 4: Build comparative models
For this step of the tutorial, all input and output files can be found in the step4_model directory of the file archive available on the index page.
Given the alignment between the template and the target TvLDH sequence, it is straightforward to build the comparative models themselves. This can be done entirely automatically using MODELLER's automodel class. It is only necessary to tell automodel where the alignment file can be found, which codes to use from the alignment for the template(s) and the target, and how many models to build.
from modeller import *
from modeller.automodel import *
env = environ()
env.io.atom_files_directory = ['.', '../atom_files']
a = automodel(env, alnfile='../step3_align/TvLDH-1y7tA.ali',
knowns='1y7tA', sequence='TvLDH', assess_methods=assess.DOPE)
a.starting_model = 1
a.ending_model = 5
a.make()
File: model-single.py
This script will run for some time, during which it should generate 5 models of the TvLDH 3D structure in PDB format named TvLDH.B99990001.pdb through TvLDH.B99990005.pdb, as well as the log file model-single.log. These PDB files can be viewed in any PDB viewer, such as Chimera.
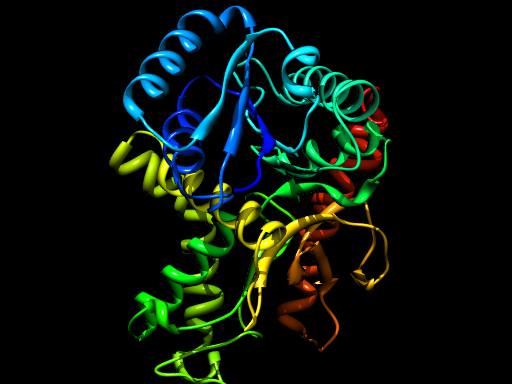
One of the generated models, rendered by Chimera and POV-Ray